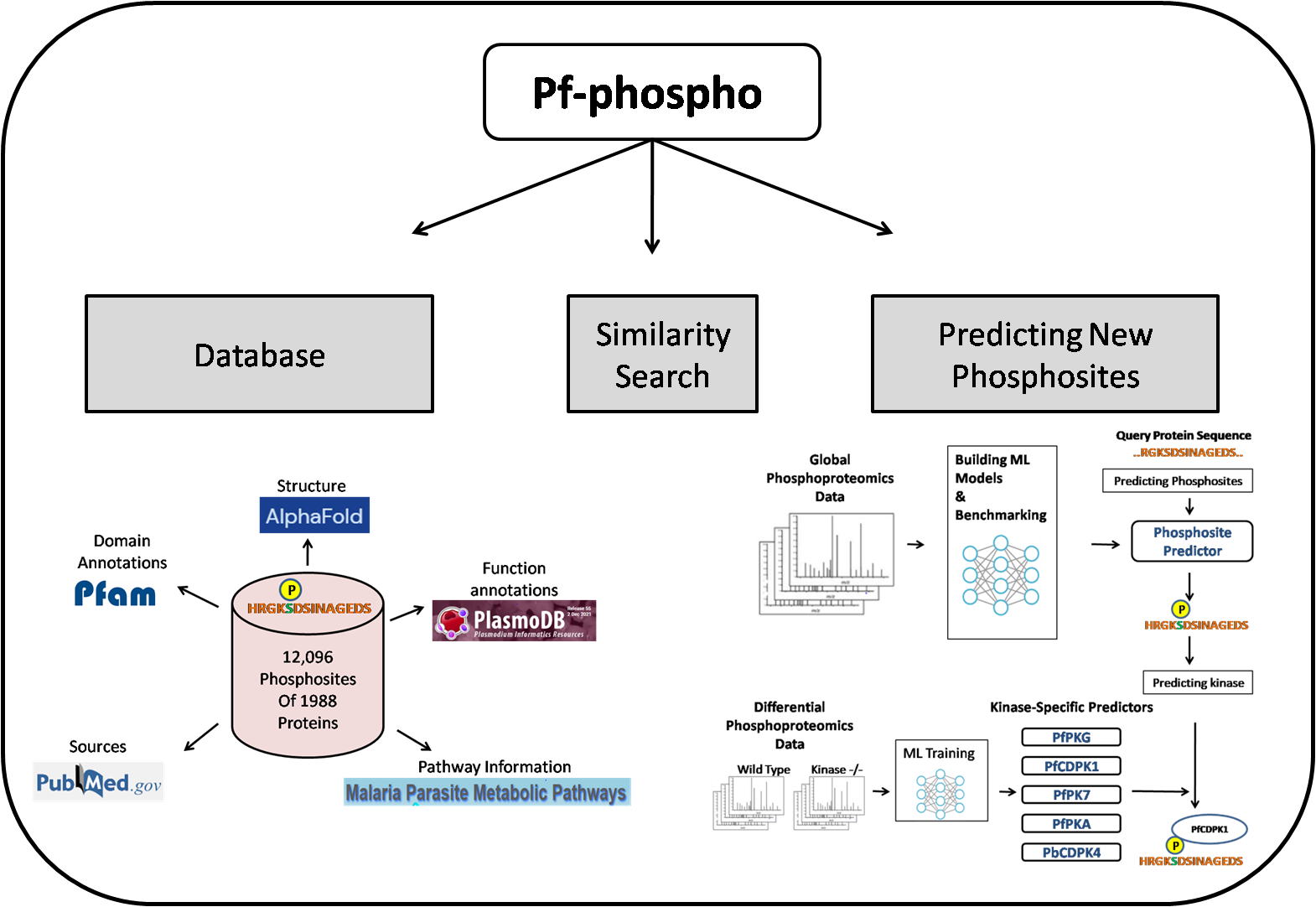
Phosphorylation is one of the pervasive post translational modifications regulating diverse cell processes and signaling. The Pf-Phospho user friendly web server caters the tools for studying phosphorylation sites of the malaria causing parasite Plasmodium. The server has two sections- the Database and the phosphosite predictor.
The Phospho-DB Database presents the well curated information of the phosphorylation sites of different proteins of Plasmodium genus species. Based on the availability, the phosphosites data has been mined from published literature studies of the mass-spectrometry based phosphosite identification experiments covering different life stages of the parasite.
The Phospho-DB Predictor is a machine learning based algorithm trained to predict new phosphosites of the Plasmodial proteins. It derives its predictive power from algorithm trained on the local sequence information and intrinsic disorderness propensity based features of the known phosphorylation sites of Plasmodium species. The predictor can predict the potential phosphorylation sites in a given protein sequence while the kinase –specific predictions can be made for 5 Plasmodial kinase- PfPKG, PfCDPK1, PfPKA, PfPK7 and PbCDPK4.