BENCHMARKING KINASE INDEPENDENT PREDICTOR
Dataset
The Kinase-independent predictor of Phospho-DB have been benchmarked on the 3098 mass-spectrometry derived phosphorylation sites of Plasmodium species published by Treeck et al., 2011, Solyakov et al., 2011, Lasonder et al., 2012, Lasonder et al., 2015 and Pease et al., 2013. The data is available at kinase_independent_predictors_data.xlsx.Results
| ROC | PR |
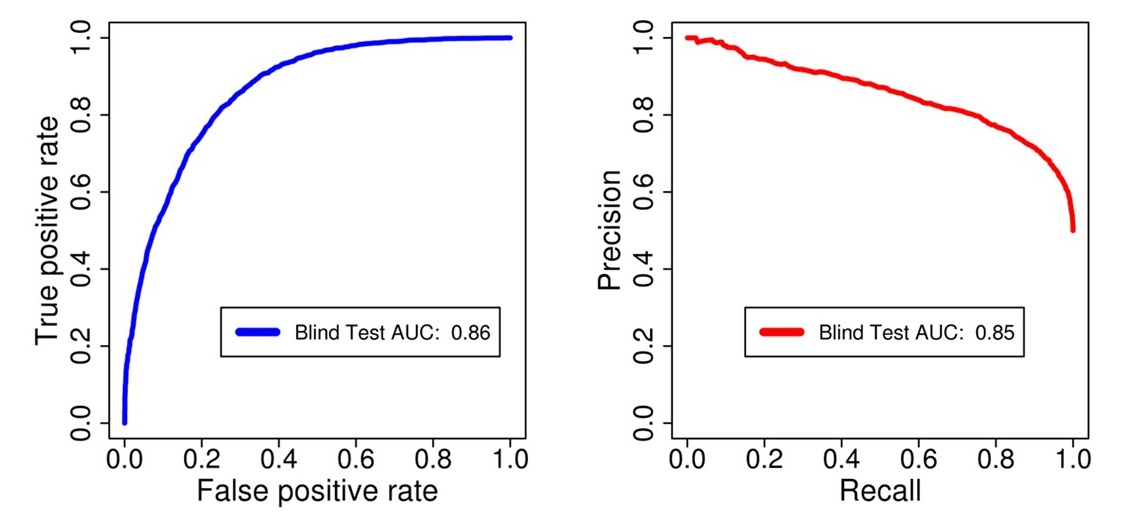
Figure: ROC and PR Curve showing the performance of kinase-independent classifier on external dataset.
Blind Testing results of the Kinase Independent Phosphosite classifier:
| Positives | Negatives | Sensitivity | FPR | Precision | F1-score | MCC | ROC AUC | PR AUC |
| 3098 | 3098 | 0.84 | 0.27 | 0.78 | 0.78 | 0.57 | 0.86 | 0.85 |
BENCHMARKING KINASE-SPECIFIC PREDICTORS
Datasets
The kinase-specific phosho-predictors for predicting phosphorylation sites of have been benchmarked with experimentally known phospho-sites of the respective kinases published by Kumar et al., 2017, Alam et al., 2015, Pease et al., 2018, Patel et al., 2019 and Invergo et al., 2017. Due to the lack of enough kinase-specific data, the Kinase-specific predicotrs have been benchmarked with only cross-validations (10-Fold). All the datasets are available at Kinase_specific_predictors_data.xlsx
Results
Performance of Kinase-specific phosphosite Predictors
| Kinase | Positives | Negatives | Sensitivity | FPR | Precision | F1-Score | MCC | ROC AUC | PR AUC |
| PfPKG | 108 | 108 | 0.8 | 0.25 | 0.77 | 0.77 | 0.55 | 0.85 | 0.84 |
| PfCDPK1 | 78 | 78 | 0.79 | 0.33 | 0.75 | 0.75 | 0.47 | 0.81 | 0.80 |
| PfPK7 | 231 | 231 | 0.84 | 0.31 | 0.77 | 0.76 | 0.53 | 0.84 | 0.83 |
| PfPKA | 60 | 60 | 0.75 | 0.33 | 0.71 | 0.71 | 0.42 | 0.79 | 0.79 |
| PbCDPK4 | 49 | 49 | 0.74 | 0.29 | 0.72 | 0.72 | 0.45 | 0.78 | 0.77 |
Comparison with performance of others Phosphosite Prediction Software
The ability of other widely used Phosphorylation site predictors – GPS5.0 (Wang et al., 2020) and NetPhorest2.1 (Miller et al., 2008) was analyzed at predicting Plasmodial phosphosites. The softwares were used to identify the known phosphorylation sites of 5 Plasmodial kinases (Kinase_specific_predictors_data.xlsx). For reference, Pf- phospho’s Kinase-specific predictors’s performance on the 10-Fold cross validation (CV) calculated on the same data set is also provided.
| Kinase Data | Predictor | Sensitivity | FPR | Precision | F1 | MCC |
|---|---|---|---|---|---|---|
| PfPKG | Pf-Phospho (CV) | 0.8 | 0.25 | 0.77 | 0.77 | 0.55 |
| GPS5.0 | 0.18 | 0.02 | 0.91 | 0.31 | 0.27 | |
| NetPhorest 2.1 | 1 | 1 | 0.5 | 0.67 | ||
| PfCDPK1 | Pf-Phospho (CV) | 0.79 | 0.33 | 0.75 | 0.75 | 0.47 |
| GPS5.0 | 0.32 | 0.05 | 0.86 | 0.46 | 0.35 | |
| NetPhorest 2.1 | 1 | 0.79 | 0.56 | 0.7 | 0.35 | |
| PfPK7 | Pf-Phospho (CV) | 0.84 | 0.31 | 0.77 | 0.76 | 0.53 |
| GPS5.0 | 0.39 | 0.25 | 0.61 | 0.48 | 0.16 | |
| NetPhorest 2.1 | 1 | 1 | 0.5 | 0.67 | ||
| PfPKA | Pf-Phospho (CV) | 0.75 | 0.33 | 0.71 | 0.71 | 0.42 |
| GPS5.0 | 0.53 | 0.33 | 0.94 | 0.68 | 0.55 | |
| NetPhorest 2.1 | 1 | 0.65 | 0.61 | 0.75 | 0.46 | |
| PbCDPK4 | Pf-Phospho (CV) | 0.74 | 0.29 | 0.72 | 0.72 | 0.45 |
| GPS5.0 | 0.43 | 0.12 | 0.78 | 0.55 | 0.34 | |
| NetPhorest 2.1 | 0.98 | 0.78 | 0.56 | 0.71 | 0.31 |